[ad_1]
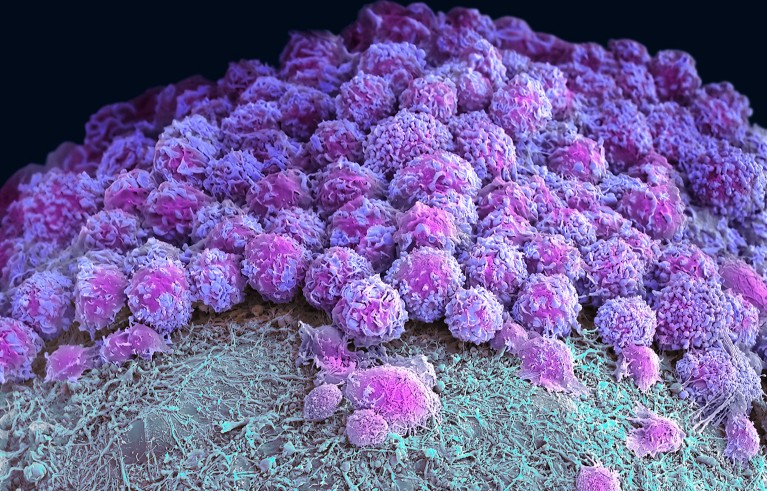
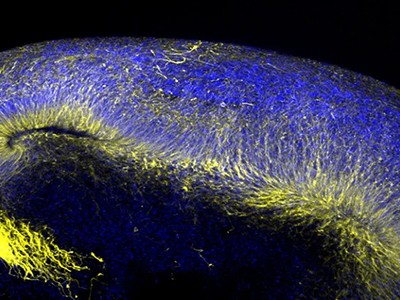
Part of a brain organoid made using human stem cells (purple).Credit: Steve Gschmeissner/Science Photo Library
Researchers have hailed organoids — 3D clusters of cells that mimic aspects of human organs — as a potential way to test drugs and even eliminate some forms of animal experimentation. Now, in two studies published on 24 April in Nature1,2, biologists have developed gut and brain organoids that could improve understanding of colon cancer and help to develop treatments for a rare neurological disorder.
“In the last ten years, people spent a lot of time to develop and understand how to make organoids,” says Shuibing Chen, a chemical biologist at Weill Cornell Medical College in New York City. “But this is the time now to think more about how to use” the models.
Organoids open fresh paths to biomedical advances
Organoids — particularly those made from human stem cells — sometimes reveal things that animal models can’t, says Sergiu Pașca, a neuroscientist at Stanford University in California and a co-author of one of the studies1. Pașca’s group studies Timothy syndrome: a genetic disorder involving autism, neurological problems and heart conditions that affects only a few dozen people in the world. Timothy syndrome is caused by a single mutation in a gene called CACNA1C, which encodes a channel through which calcium ions enter cells including neurons.
Pașca says that there are no good animal models for Timothy syndrome because the underlying mutation doesn’t always cause the same symptoms in rodents. “It became very clear to us we’d need to find a way of testing in vivo,” he says.
Cultured cells
The researchers turned to brain organoids to recreate the disorder. They took stem cells from 3 people with the mutation that causes Timothy syndrome and cultured them for about 250 days, treating the cells with signalling molecules that encouraged them to turn into brain organoids containing every type of neuron found in the cerebral cortex, the outer layer of the brain. To create a more lifelike environment for the organoids, the team then injected these structures into the brains of rats, where the cells formed connections with the rodents’ own neurons. This made a system in which the researchers could test potential treatments for the disorder.
Human neurons have four different forms of this calcium channel, but only one of them is defective in Timothy syndrome. Getting rid of the mutated channel, the researchers suggest, would allow the other, healthy channels to take over.
Organoids grown from amniotic fluid could shed light on rare diseases
To do this, they identified short pieces of nucleic acids called oligonucleotides that can stop cells producing the mutated form of the protein by interfering with genetic transcripts. Around two weeks after the researchers injected these oligonucleotides into the rats’ brains, most of the defective calcium channels in the organoids and the surrounding rat neurons had been replaced by other versions of the protein. The neurons in the organoids had also changed shape — from small, less complex forms similar to those in people with Timothy syndrome to larger, more complex shapes typical of healthy neurons. “To be honest, I didn’t think it would work so well,” Pașca says.
He adds that his group hope to test the therapy on people in clinical trials eventually, although they will first need to prove that the oligonucleotides are safe by testing them in non-human primates. The researchers think that the treatment would be effective for about three months, so people would need to receive frequent injections. The advantage, Pașca says, is that the biological effects of the treatment would be reversible and any side effects would be short-lived.
Miniature colon
In a separate paper2, bioengineer Matthias Lütolf at the Swiss Federal Institute of Technology in Lausanne and his colleagues used mouse stem cells to model tissue from the other end of the body: the gut tissue that makes up the colon and rectum. Organoids tend to grow in tight balls, so the researchers grew the cells on a scaffold to recreate the tube structures seen in real gut tissue.
To make a model of colon cancer, they engineered the cells to contain light-sensitive proteins attached to cancer-causing genes. This allowed them to use a blue laser to switch on the genes and trigger the growth of tumours at specific sites in the organoid, then watch how the tumours changed over the course of weeks.
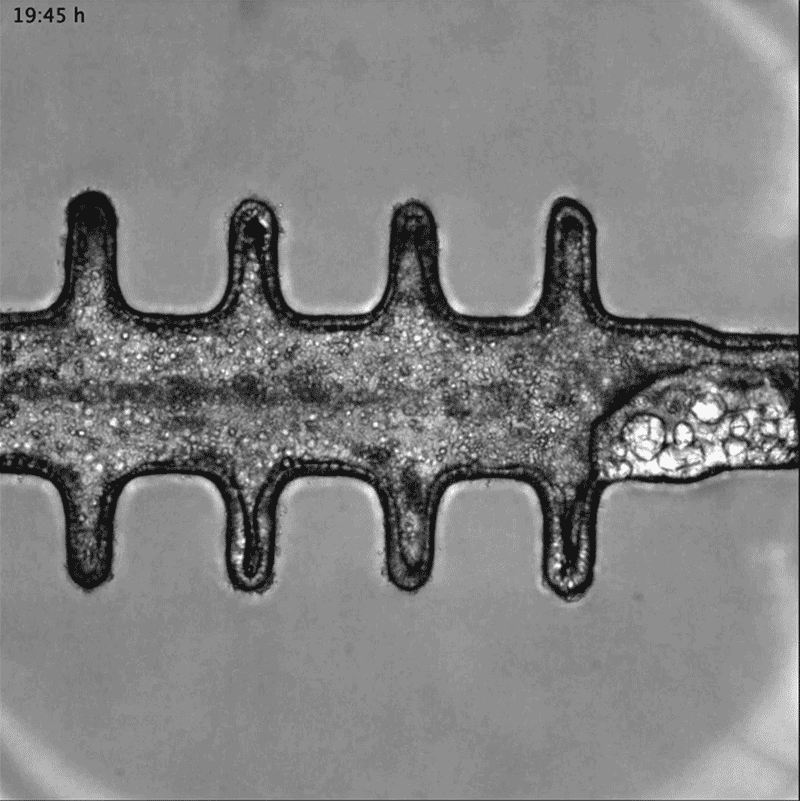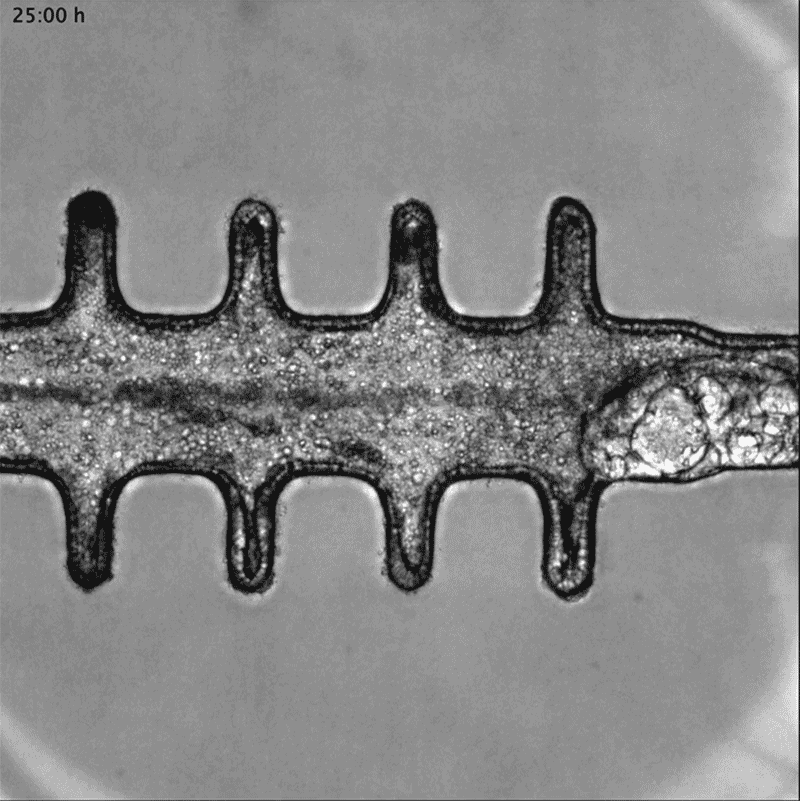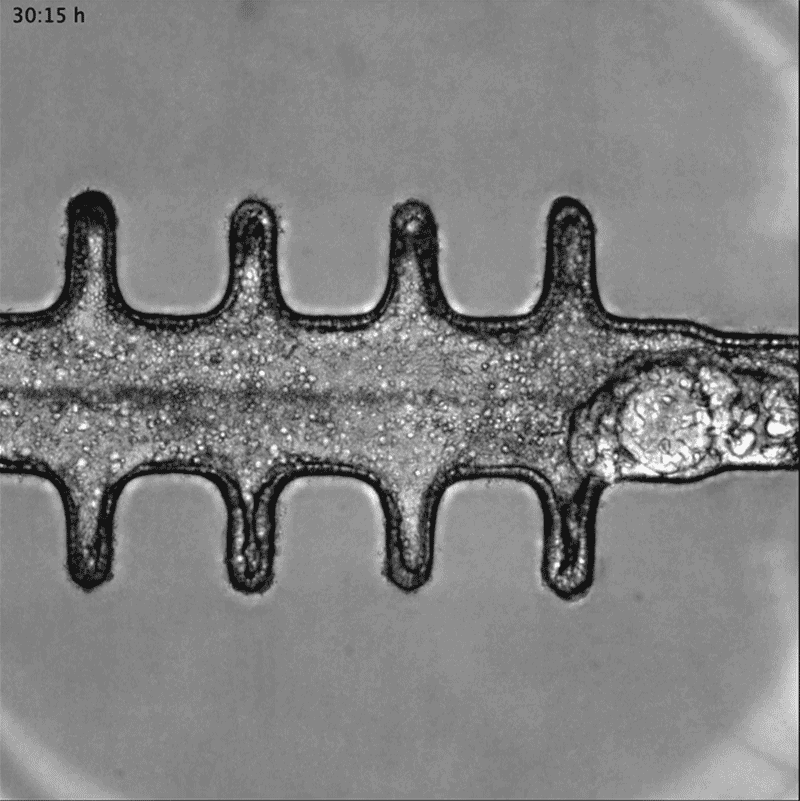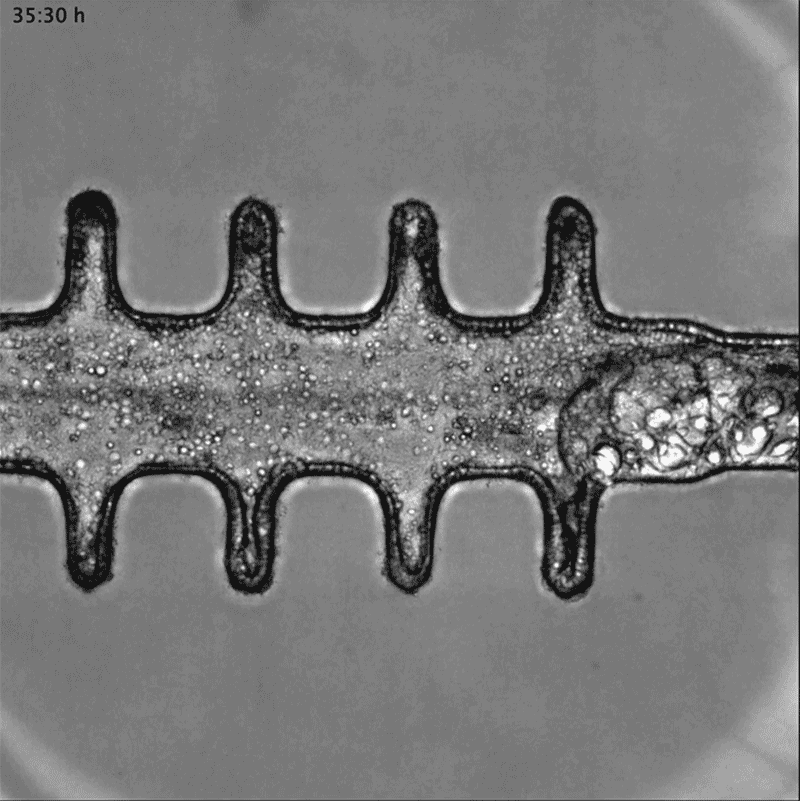
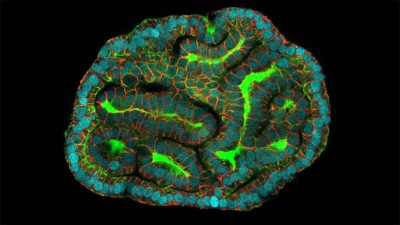
Researchers grew tumours inside a model of gut tissue created using mouse stem cells.Credit: Matthias P. Lutolf et al/Nature
When the researchers injected the cancerous cells into mice, the tumours looked similar to those seen in human colorectal cancer. The organoids accumulated fewer tumours when the researchers restricted calories in their medium, which also happens in people with colorectal cancer.
Lütolf was struck by the difference between the tumours triggered throughout the organoid. It would be difficult, he says, to use the organoids to screen a large number of new drugs, because of the differences and because they take a long time to produce. But he says organoids could be useful in investigating how drugs or immunotherapies kill tumours, and how factors such as a person’s environment and immune system affect the development and progression of colorectal cancer.
The mini lungs and other organoids helping to beat COVID
Lütolf and his group plan to manipulate the gut organoids to better reflect the human system, and hope that they could eventually replace animal models in some instances. Future organoid models, Lütolf says, could include bacteria that live in the gut or could be exposed to different levels of oxygen that mimic those available in various anatomical regions of the gut.
The two studies are “very well-designed”, Chen says. She adds that the organoid models seem particularly useful in showing the complexity of diseases such as Timothy syndrome and colorectal cancer, and how they progress over time. Now that researchers have found good ways to make organoids that model different organs, she says, the next steps will be learning how to scale up production for drug development, and making them more complex so that they reflect real human biology.
[ad_2]
Source Article Link