[ad_1]
Antibody and antigen production
We experimentally validated the 376 designed candidates. To leverage available resources at multiple experimental sites, we split candidates into partially overlapping sets 1 and 2. Set 1 consisted of 230 designs expressed as IgG in HEK-293 cells (ATUM), and set 2 consisted of 204 designs expressed as IgG via a pVVC-mCisK_hG1 vector (Twist BioScience) in transiently transfected CHO cells. Omicron antigens were produced in Expi293F cells (Thermo Fisher Scientific) and purified on HisTrap Excel columns (Cytiva).
In the following experiments, we selected antigens or viral strains to gauge the success of three goals: (1) improving binding affinity and efficacy to BA.1 and BA.1.1; (2) maintaining efficacy to historical strains, for which design explicitly targeted Delta but experiments often substituted WA1/2020 D614G; and (3) determining robustness to emerging VOCs.
Designed antibodies maintain expression
Because in silico derivatization of antibody sequences can compromise production yield, we measured the concentrations of the 230 COV2-2130-derived recombinant antibodies in set 1 and compared these concentrations to that of the parental antibody. The purified concentrations of 73.9% of redesigned antibodies exceeded that of the parental COV2-2130 antibody (170 of 230 monoclonal antibodies at more than 171.2 mg l−1), reaching as high as 305 mg l−1. Our designed candidates for downstream characterization retained fundamental production properties of the parental antibody, with just 10% of designed antibodies producing poor yields relative to the parental molecule (22 of 230 monoclonal antibodies at less than 135 mg l−1, that is, less than 80% of the parental antibody yield).
Thermostability and binding Omicron
We screened all designed antibodies for binding to RBDs. Set 1 was screened via a single-concentration immunoassay (Gyrolab xPlore) in the contexts of WA1/2020, Delta, BA.1 or BA.1.1 RBDs (Extended Data Fig. 1). For set 2, we used a multi-concentration immunoassay (ELISA; Extended Data Fig. 2) in the context of wild-type, BA.1 or BA.1.1 RBDs. In the single-concentration immunoassay, this value was chosen as a single dilution factor, causing most designed antibody samples to fall in the dynamic range of the positive control. In both cases, we compared the binding of the designed antibodies with a broadly cross-reactive, non-ACE2-competitive control antibody (S309)24 and the parental COV2-2130 antibody. As intended, most antibody designs had altered binding profiles, indicating that the designed mutations were consequential. Approximately 11% of the designs of set 1 retained WA1/2020 antigen binding at the measured concentration; roughly 6% improved binding to BA.1, and 5% did so to BA.1.1. The corresponding numbers for set 2 were 9% to WA1/2020 and 8% to BA.1. Following this initial screen, we downselected both sets of antibody designs to those with improved binding to Omicron subvariants BA.1 and BA.1.1 for further characterization.
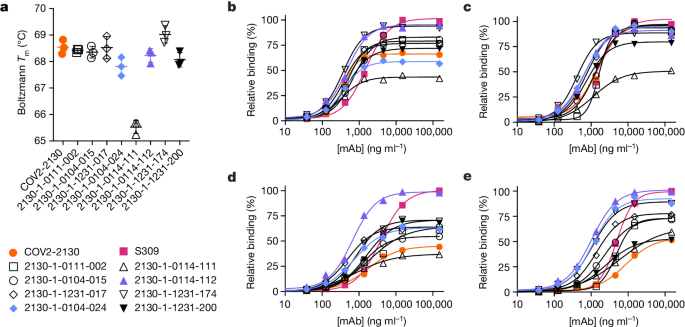
These downselected antibodies were re-manufactured at larger scale. We characterized the resulting IgG antibodies by immunoassay and thermal shift (melt temperature) assessments. In agreement with our screens, seven of the eight top-performing antibodies preserved comparable binding with WA1/2020 and Delta RBDs, improving over the parental COV2-2130 antibody with respect to their binding to Omicron BA.1 and BA.1.1 RBDs (Fig. 2). Furthermore, seven of the eight antibodies had melting temperatures and expression properties comparable with those of COV2-2130. One antibody, 2130-1-0114-111, had reduced melting temperature (Extended Data Table 1). The antibody 2130-1-0114-112 displayed best-in-class binding across all RBD variants and had no substantial difference in thermal stability compared with the parental COV2-2130 antibody.
a, The parental COV2-2130 (orange circles) and computationally designed antibodies (2130-1-0114-112 in purple triangles, 2130-1-0104-024 in blue diamonds and remainder in black) were assayed for thermal shift (n = 3 technical replicates). Melting temperature (Tm ) calculated based on the Boltzmann method. Data are mean and s.d. b–e, The parental COV2-2130 antibody and computationally designed antibodies (see symbols in a) and cross-reactive positive control antibody S309 (magenta squares) were analysed for relative binding to four SARS-CoV-2 spike RBD variants in the Gyrolab immunoassay: WA1/2020 (b), Delta B.1.617.2 (c), Omicron BA.1 (d) and Omicron BA.1.1 (e). Lines represent a four-parameter logistic regression model fit using GraphPad Prism to each titration, executed without technical replicates. mAb, monoclonal antibody.
Restored pseudoviral neutralization
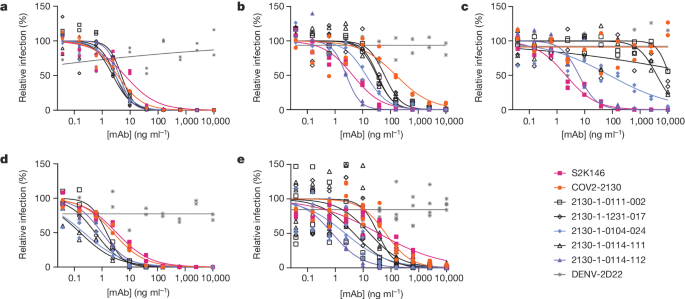
We performed pseudovirus neutralization assays to characterize the functional performance of five selected antibody designs (Fig. 3 and Extended Data Table 2), compared with parental COV2-2130; the positive control S2K146 (ref. 25), which competes with ACE2 binding; and the negative control DENV-2D22 (ref. 26). Our designs maintained neutralization activity against pseudoviruses displaying historical spike proteins (WA1/2020 D614G) and achieved neutralization of those with Omicron BA.1 spikes. The single-best candidate design, 2130-1-0114-112, restored potent neutralization in the context of BA.1.1 and showed a two-order-of-magnitude improvement in the half-maximal inhibitory concentration (IC50) versus parental COV2-2130 for BA.1 and BA.4. These pseudovirus neutralization test results showed that our designs neutralized BA.2 and BA.4 more potently than COV2-2130, despite the emergence of these VOCs after the conception of our designs.
a–e, The parental COV2-2130 antibody (orange circles), the cross-reactive positive control antibody S2K146 (magenta squares), the negative control antibody DENV-2D22 (grey x) and down-selected computationally designed antibodies (symbols as indicated in the key) were assayed by neutralization with lentiviruses pseudotyped with spike variants of WA1/2020 D614G (a), Omicron BA.1 (b), Omicron BA.1.1 (c), Omicron BA.2 (d) and Omicron BA.4 (e). Curves are four-parameter logistic regression models fit to two (a–d) or four (e) replicate serial dilutions using GraphPad Prism.
Restored authentic virus neutralization
We evaluated 2130-1-0114-112 (containing four mutations: GH112E, SL32A, SL33A and TL59E) for authentic virus neutralization performance against several strains of SARS-CoV-2 by a focus reduction neutralization test in Vero-TMPRSS2 cells (Extended Data Fig. 3). The strains that we used included several Omicron targets: WA1/2020 D614G, Delta (B.1.617.2), BA.1, BA.1.1, BA.2, BA.2.12.1, BA.4, BA.5 and BA.5.5. In all cases apart from Delta, 2130-1-0114-112 had an IC50 < 10 ng ml−1. Compared with the parental COV2-2130, 2130-1-0114-112 restored potent neutralization activity to both BA.1 (8.08 ng ml−1) and BA.1.1 (7.77 ng ml−1), showed a more than fivefold improvement in IC50 to BA.2 (2.4 ng ml−1) and BA.2.12.1 (2.27 ng ml−1), and conferred 50-fold or greater improvements in IC50 to BA.4 (3.16 ng ml−1), BA.5 (3.51 ng ml−1) and BA.5.5 (5.29 ng ml−1). We also evaluated 2130-1-0114-112 and a less-mutated alternative design, 2130-1-0104-024 (containing two mutations: SL32W and TL59E), in plaque assays with Vero E6-TMPRSS2-T2A-ACE2 cells (Extended Data Fig. 4). IC50 values for 2130-1-0104-024 were 37.7 ng ml−1, 75.94 ng ml−1 and 781.7 ng ml−1 for Delta, BA.1 and BA.1.1 viruses, respectively.
Prophylaxis in vivo
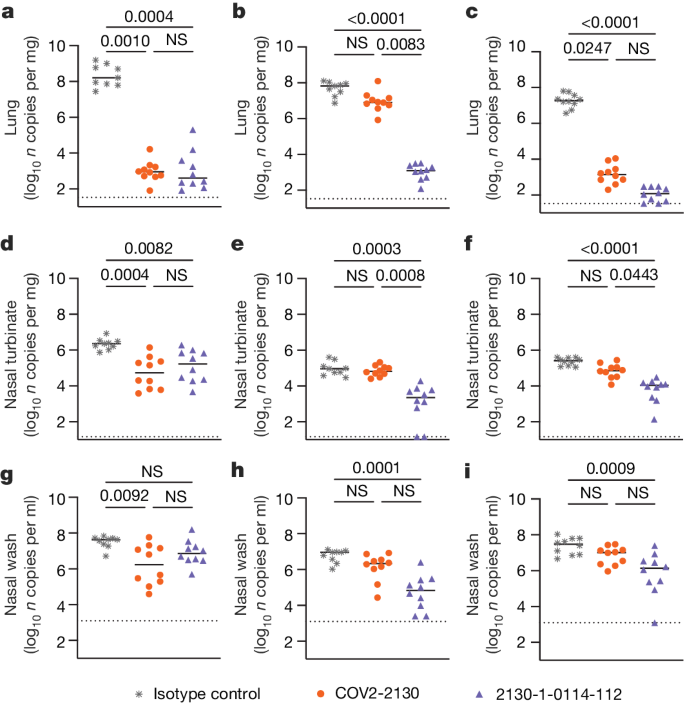
To compare the prophylactic efficacy of 2130-1-0114-112 and the parental COV2-2130 monoclonal antibody in vivo, we administered a single 100 μg (approximately 5 mg kg−1 total) dose to K18-hACE2 transgenic mice 1 day before intranasal inoculation with WA1/2020 D614G, BA.1.1 or BA.5 (88 mice in total, 9–10 for each monoclonal antibody and viral strain). Although Omicron viruses are less pathogenic in mice than in humans, they replicate efficiently in the lungs of K18-hACE2 mice27,28. Viral RNA levels were measured at 4 days post-infection in the nasal washes, nasal turbinates and lungs (Fig. 4). As expected, the parental COV2-2130 monoclonal antibody effectively reduced WA1/2020 D614G infection in the lungs (180,930-fold), nasal turbinates (42-fold) and nasal washes (25-fold) compared with the isotype control monoclonal antibody. However, the COV2-2130 monoclonal antibody lost protective activity to BA.1.1 in all respiratory tract tissues, whereas to BA.5, protection was maintained in the lungs (13,622-fold) but not in the nasal turbinates or nasal washes. Compared with the isotype control monoclonal antibody (Fig. 4), 2130-1-0114-112 protected against lung infection by WA1/2020 D614G (399,945-fold reduction), BA.1.1 (53,468-fold reduction) and BA.5 (160,133-fold reduction). Moreover, in the upper respiratory tract, 2130-1-0114-112 also conferred protection to WA1/2020 D614G, BA.1.1 and BA.5. The differences in protection between the parental COV2-2130 and derivative 2130-1-0114-112 monoclonal antibodies were most apparent in mice infected with BA.1.1, which directly parallels the neutralization data (Fig. 3 and Extended Data Figs. 3 and 4).
a–i, Eight-week-old female K18-hACE2 mice were administered 100 μg (approximately 5 mg kg−1) of the indicated monoclonal antibody treatment by intraperitoneal injection 1 day before intranasal inoculation with 104 focus-forming units (FFU) of WA1/2020 D614G (a,d,g), Omicron BA.1.1 (b,e,h) or Omicron BA.5 (c,f,i). Tissues were collected 4 days after inoculation. Viral RNA levels in the lungs (a–c), nasal turbinates (d–f) and nasal washes (g–i) were determined by RT–qPCR (lines indicate median of log10 values); n = 9 (WA1/2020 D614G and BA.1.1 isotype control groups) or 10 (all others) mice per group, from two experiments. The limit of assay detection is shown as a horizontal dotted line. Statistical comparisons between groups were by Kruskal–Wallis ANOVA with Dunn’s multiple comparisons post-test; P values are as listed or not significant (NS) if P > 0.05. All analyses were conducted in GraphPad Prism.
Potency without additional liabilities
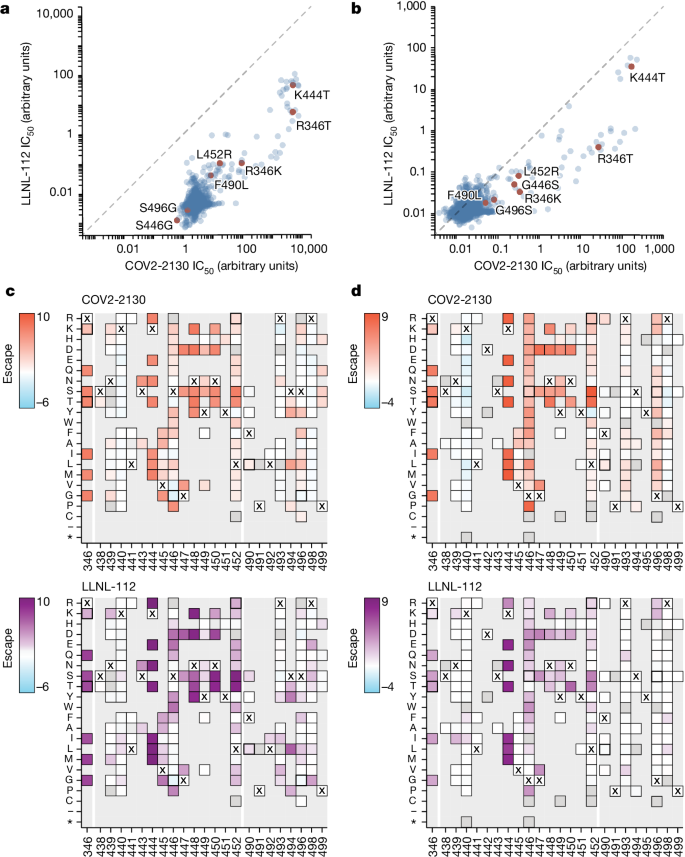
To understand the neutralization breadth of 2130-1-0114-112 relative to its ancestral antibody, we mapped the epitopes for both antibodies using spike-pseudotyped lentiviral deep mutational scanning (DMS)29. For each antibody, we mapped escape mutations in both the BA.1 and the BA.2 spikes. DMS experiments showed that the escape profile of both COV2-2130 and 2130-1-0114-112 in the context of both BA.1 and BA.2 backgrounds is consistent with the epitope of the antibodies, but with differences in sensitivity to particular mutations (Fig. 5). Consistent with live and pseudovirus neutralization assays (Fig. 3 and Extended Data Figs. 3 and 4), mutations at RBD positions R346 and L452 are sites of substantial escape from both antibodies (Fig. 5). In addition, both antibodies lose potency with mutations at site K444 (such as K444T found in BQ.1* variants). The reversion mutation S446G in the BA.1 background increases the neutralization potency of both antibodies (negative escape values in heatmaps) (Fig. 5c) and probably contributes to greater neutralization potency to the BA.2 variant (Fig. 3 and Extended Data Fig. 3), which carries G446. Most mutations at RBD sites K440 and R498 are slightly sensitizing to the COV2-2130 antibody in both BA.1 and BA.2 backgrounds, but provide weak escape for 2130-1-0114-112 in the BA.1 background and have even weaker effect in the BA.2 background. In agreement with pseudovirus neutralization assays (Fig. 3), comparison of mutation-level escape showed that the 2130-1-0114-112 antibody is substantially more potent than COV2-2130 to the BA.1 variant and retains better potency against viruses with additional mutations in both BA.1 and BA.2 backgrounds (Fig. 5a,b). However, even with improved potency, 2130-1-0114-112 is still vulnerable to escape at multiple RBD residues in the 444–452 loop, which is the site of convergent substitutions in several Omicron lineages30. Many of these variants contain multiple substitutions at positions identified by DMS as important for neutralization or in close proximity to the COV2-2130 epitope, including BQ.1.1 (R346T and K444T), XBB (R346T, V445P and G446S) and BN.1 (R346T, K356T and G446S). To understand the impact of these VOCs, we assessed the ability of 2130-1-0114-112 to neutralize BQ.1.1, XBB and BN.1 in pseudoviral neutralization studies. Consistent with the previously known liabilities of COV2-2130 and our DMS results, 2130-1-0114-112 loses neutralizing activity to these VOCs (Extended Data Fig. 5), probably due to substitutions at 444 and combinatorial effects of multiple substitutions within the COV2-2130 epitope present in these variants. Together, these data demonstrate that 2130-1-0114-112 exhibits improved potency against many individual substitutions without incurring additional escape liabilities, although RBD residues such as 444 remain critical for neutralization activity of both 2130-1-0144-112 and COV2-2130.
a,b, Comparison between IC50 values measured using DMS for COV-2130 and 2130-1-0114-112 antibodies in BA.1 (a) and BA.2 (b) backgrounds, with key mutations highlighted. Arbitrary units in both plots are on the same scale. Interactive plots that display each mutation can be found at https://dms-vep.org/SARS-CoV-2_Omicron_BA.1_spike_DMS_COV2-2130/compare_IC50s.html for the BA.1 background and at https://dms-vep.org/SARS-CoV-2_Omicron_BA.2_spike_DMS_COV2-2130/compare_IC50s.html for the BA.2 background. c,d, Heatmaps of mutation escape scores at key sites for each antibody in BA.1 (c) and BA.2 (d) backgrounds. Escape scores were calculated relative to the wild-type amino acid in the same virus background. X marks wild-type amino acid in the relevant background. Amino acids not present in the DMS libraries lack squares; grey squares are mutations that strongly impair spike-mediated infection. Mutations identified in a,b are shown with a heavy line surrounding the corresponding box in c,d. Interactive heatmaps for full spike can be found for the BA.1 background at https://dms-vep.org/SARS-CoV-2_Omicron_BA.1_spike_DMS_COV2-2130/COV2-2130_vs_2130-1-0114-112_escape.html and https://dms-vep.org/SARS-CoV-2_Omicron_BA.2_spike_DMS_COV2-2130/COV2-2130_vs_2130-1-0114-112_escape.html for the BA.2 background.
Structural basis for restored potency
To elucidate the key intermolecular interactions that form the interface and determine Omicron RBD recognition by 2130-1-0114-112, we performed 3D reconstructions of the complex between the SARS-CoV-2 Omicron BA.2 spike and the 2130-1-0114-112 Fab fragment using cryo-electron microscopy (cryo-EM). Reconstruction using refinement of the full complex gave a map with average resolution of 3.26 Å, but the interface region between the BA.2 RBD and the 2130-1-0114-112 Fab was not well resolved, presumably due to the flexibility of the RBD–Fab region in the reconstruction. To resolve details at the intermolecular interface, we performed focused refinement of this portion of the structure. Focused refinement resulted in an effective resolution of approximately 3.6 Å for this region (Electron Microscopy Data Bank EMD-28198 and EMD-28199, and Protein Data Bank 8EKD) (Fig. 6 and Extended Data Fig. 6).
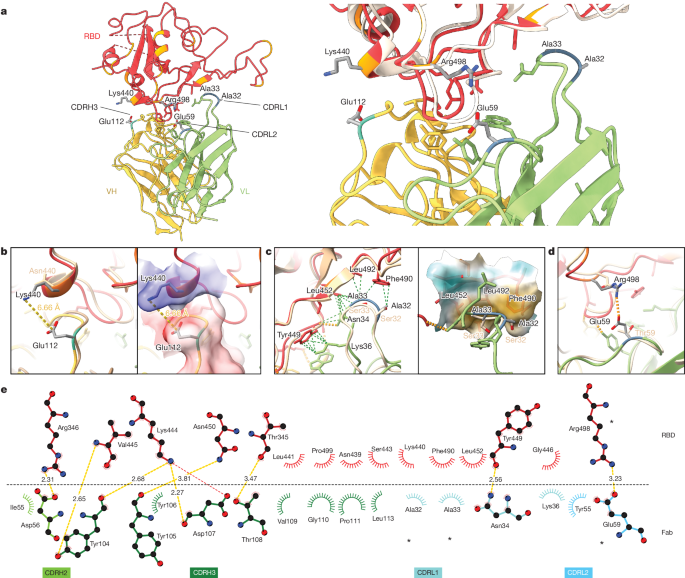
a, Atomic model of the RBD–Fab complex, coloured by chain: BA.2 RBD in red, 2130-1-0114-112 HC in yellow and 2130-1-0114-112 LC in green. BA.2 RBD mutations are in orange, and 2130-1-0114-112 mutations are in cyan and blue (HC and LC) (left). A close-up view of the RBD–Fab interface, showing WA1 RBD (Protein Data Bank 7L7E, light brown shading) aligned with the BA.2 RBD (right). b–d, Details showing the 2130-1-0114-112 modified residues and their interaction with BA.2 RBD, coloured as in a. Residue labels are shown in black for the BA.2 complex and brown for the overlaid WA1-2130 complex. The orange and green dashed lines indicate hydrogen bond and hydrophobic interactions, respectively; the yellow dashed lines are labelled with distances. CDRH3 residue Glu112 (left) and with the surface coloured by electrostatic potential (right), showing the positive and negative charges of RBD Lys444 and CDRH3 Glu112 (b). CDRL1 Ala32 and Ala33 hydrophobic network (left) and with the nearby RBD surface coloured by hydrophobicity (right; orange to cyan indicates hydrophobic to hydrophilic) (c). CDRL2 Glu59 salt bridge with RBD residue Arg498 (d). e, 2D diagram of Fab 2130-1-0114-112 paratope and epitope residues involved in hydrogen bonds and salt bridges (yellow and red dashed lines, respectively; distances in Å) and hydrophobic interactions (curved lines with rays). Atoms are shown as circles, with oxygen, carbon and nitrogen in red, black and blue, respectively. Interacting residues that belong to CDR loops are coloured in corresponding shades. The asterisks indicate mutated residues. Image created with Ligplot+34.
This model shows the binding interface of 2130-1-0114-112–RBD and elucidates how 2130-1-0114-112 regains neutralization potency to Omicron VOCs. The parental COV2-2130 forms extensive interactions with the RBD through CDRH2 and CDRH3, as well as CDRL1 and CDRL2 (ref. 13) with hydrogen bond networks and hydrophobic interactions. To improve binding interactions with Omicron subvariants, 2130-1-0114-112 modifies three CDR loops: G112E in CDRH3, S32A and S33A in CDRL1, and T59E in CDRL2.
The RBD N440K substitution, identified in the DMS as sensitizing for escape from COV2-2130 but less so for 2130-1-0114-112, is on the edge of the interface with the 2130-1-0114-112 CDRH3 loop and does not make direct contact with the CDRH3 substitution G112E. However, N440K introduces a positive charge to a local environment that has substantial hydrophobic-to-hydrophobic contact. The negative charge introduced by the CDRH3 G112E substitution (Fig. 6b) might improve the electrostatic interactions in this region. It is possible that E112 and K440 are interacting by coordinating a water molecule, but the structural resolution is not sufficient to confirm this type of interaction. These experimental structural results are also consistent with our molecular dynamics simulations, which showed this transient interaction between CDRH3 E112 and RBD K440.
The local environment around the CDRL1 loop is mostly hydrophobic (comprising the RBD residues L452, F490 and L492, as well as the Omicron mutation E484A) with a hydrogen bond from LC N34 (Fig. 6c). The hydrophilic-to-hydrophobic CDRL1 substitutions introduced in 2130-1-0114-112, S32A and S33A, may favour the local environment and strengthen hydrophobic interactions with the RBD (Fig. 6c,e). This is supported by the DMS identification of sensitivity to hydrophobic-to-hydrophilic substitutions at RBD position 452 for both 1230-1-0114-112 and the parental COV2-2130. Finally, the T59E mutation in the CDRL2 loop establishes a new salt bridge with the RBD substitution Q498R present in Omicron RBDs. This new salt bridge probably strengthens the interaction with the RBD (Fig. 6d,e).
2130-1-0114-112 distributes four substitutions across three of the four CDR loops comprising the parental COV2-2130 paratope. Mutations to CDRH3 loop were less fruitful than mutations in the L1 and L2 (Extended Data Fig. 7a compared with Extended Data Fig. 7d) when looking across all antibody candidates. Among successful candidates, substitutions at positions 32 and 33 in CDRL1 appear enriched—particularly with hydrophobic residues—consistent with our analysis of this region of the experimentally solved structure of 2130-1-0114-112–BA.2 spike. Another candidate, 2130-1-0104-024, achieves improved affinity and neutralization with only two substitutions: S32W in CDRL1 and T59E in CDRL2. However, full neutralization potency is not reached without the potential charge accommodation mediated by G112E. This suggests that a combination of new bonds and accommodating charge changes optimized the restored affinity and potency of 2130-1-0114-112 with Omicron variants (Extended Data Fig. 8). Altogether, the structural model of 2130-1-0114-112 with the BA.2 RBD helps explain the observed restoration of potency to early SARS-CoV-2 Omicron VOCs.
[ad_2]
Source Article Link