[ad_1]
Scientists have used microorganisms to produce beneficial chemicals for decades. By providing the microbes with enzymes and metabolic pathways, researchers can coax cells to churn out everything from food additives to biofuels.
One advantage of biomanufacturing is ecological: the processes are generally more environmentally friendly than are chemical manufacturing methods. But it’s expensive, mainly because cells won’t create something simply because researchers want them to. The enzymes needed to produce ethanol fuel from plant sugars, for example, must compete with the cell’s own metabolic enzymes for energy, and the yield can be poor.
Building complex products, such as pharmaceuticals, can involve numerous steps carried out by multiple enzymes. When the cell’s ability to develop is pitted against its ability to make a product it doesn’t need, “the cells are going to choose to grow every time”, says Brian Pfleger, a synthetic biologist at the University of Wisconsin–Madison.
Rather than attempting to win this metabolic-resource war, some researchers are trying to sidestep it, introducing parallel biosynthesis pathways that won’t interfere with natural processes. They’ve zeroed in on cofactors: small organic molecules that bind to enzymes and donate (reduce) or accept (oxidize) the electrons needed for enzymes to do their work. Many of these ‘redox’ reactions, in which chemical groups are added, subtracted or restructured in compounds, are energetically expensive for cells. Compounding the problem is the fact that some reactions require a relatively ‘reducing’ environment whereas others require an ‘oxidizing’ environment; making both conditions occur in the same cell is nearly impossible.
Designing synthetic redox cofactors that can be used only by synthetic enzymes could bypass the cell’s natural machinery entirely, ensuring that the two processes no longer need to compete. “It’s still using nature’s same infrastructure,” says Han Li, a chemical and biological engineer at the University of California, Irvine. These non-canonical redox cofactors (NRCs), she says, could allow synthetic biologists to create chemical reactions that are either entirely new to nature, or more efficient than those catalysed by existing enzymes.
Li’s group is among several that are engineering NRCs, and the enzymes to use them. They have already designed synthetic systems that yield more product than previous attempts. But it isn’t easy. Researchers need to find ways to efficiently produce NRCs, ensure that the cofactors and enzymes don’t harm the cells, and modify each enzyme to accept NRCs without disrupting the enzyme’s normal functions.
Scaling up NRC production to industrial levels will bring more challenges. But some researchers think that NRCs could transform chemical manufacturing, making bioproduction cheaper, easier and more viable. “Right now, biomanufacturing can’t compete with fossil-fuel based processes because of cost,” Li says. By decreasing the cost, she says, waste and emissions could decrease as well.
A universal currency
The idea of fleshing out biomanufacturing processes with a parallel set of cofactors has been around since at least the 1950s, when researchers began finding ways to synthesize alternative compounds and searching for the rare enzyme that could use them effectively. But the artificial cofactors were prohibitively expensive to make. And because natural enzymes could switch back and forth between synthetic and natural cofactors, both the cell and biomanufacturing yields suffered. Designing enzymes that specifically used a non-canonical cofactor proved an even harder problem, until advances in genetics and structural biology made protein engineering a relatively straightforward process.
Li was inspired by efforts to produce synthetic nucleotides that could be inserted into DNA, allowing genetic sequences to use six different nucleotides instead of four and drastically expanding the number of amino acids that the DNA could theoretically encode. If the same idea could be applied to cellular metabolism, she reasoned, it would expand the types of product that cells could make. “Why not do this for the universal currency of life?” she says.
How scientists are hacking the genetic code to give proteins new powers
Cofactors are indeed a universal currency. Plant, animal and microbial cells all tend to use the same organic molecules to drive enzymatic reactions, such as ATP for energy and S-adenosyl methionine (SAM) as a source of methyl and sulfur-containing chemical groups.
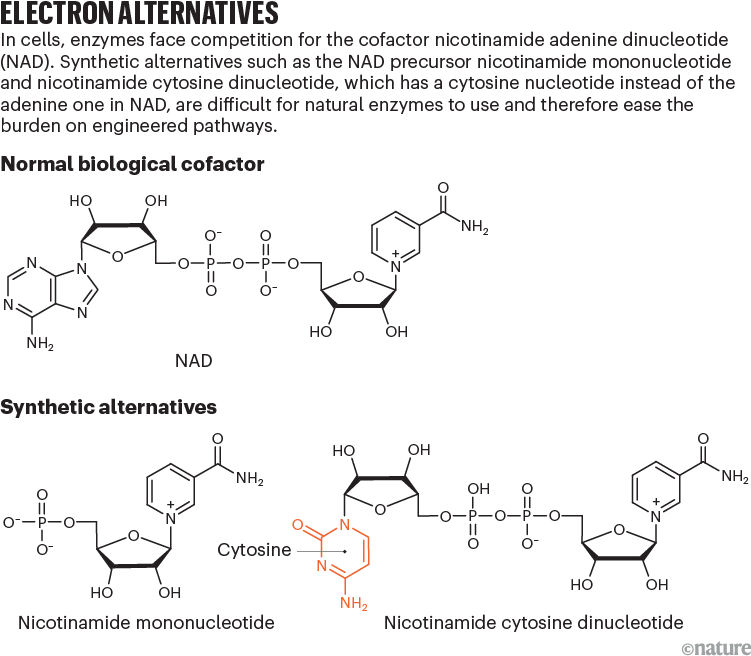
One of the most common cofactors is nicotinamide adenine dinucleotide (NAD), which, with its phosphorylated form NADP, drives metabolic pathways, such as those that break down glucose into carbon dioxide and water. These cofactors exist in two configurations: a reduced form (NADH) that donates electrons and an oxidized form (NAD+) that receives them.
A genomic analysis of the bacterium Escherichia coli estimated that the organism uses NAD in 127 enzymatic reactions and NADP in another 1131. NAD “is so central to life and all the ways that we understand it”, concludes Ruud Weusthuis, a microbial engineer at Wageningen University in the Netherlands.
But that centrality also means that, when a bioengineer wants to coax microbes into producing a new product — an artificial scent, for instance — they face a lot of competition for NAD. Simply providing the cells with more cofactors doesn’t help because the cells’ metabolisms just run faster. And given the choice, natural enzymes prefer to carry out easier reactions that don’t require much energy, making it hard for engineers to restrict where electrons are used. “Electrons are important, and we don’t want to waste them,” Weusthuis says.
NRCs can shift that balance of power. One of the most promising is nicotinamide mononucleotide (NMN), a subunit of NAD (see ‘Electron alternatives’). Bacterial cells naturally synthesize small amounts of NMN when producing the nucleotides that make up DNA and RNA, and adding certain enzymes to the cells can increase production.
In a 2019 paper2, Li’s team made four changes to an enzyme called glucose dehydrogenase that made it 10 million times more likely to use NMN than NAD. By pairing this modified enzyme with another that can naturally use NMN, the group successfully induced E. coli to produce a pharmaceutical product called levodione using only artificial cofactors. The swap produced about as much levodione as the cells could produce with enzymes that use only NAD, and altering the enzyme’s structure further could increase its efficiency, the researchers say.
Li’s team has also used NMN to power more challenging syntheses, including the scent-related chemical citronellal. Citronellal is difficult to make because cells quickly convert it into other products. Engineering certain enzymes to use NMN exclusively instead of NAD allowed citronellal to accumulate in cells3. But, in a finding that highlights the delicate balance metabolic engineers face, Li’s group also found that when NMN+ concentrations are high, cell growth suffers. Bioengineer William Black, who works with Li at the University of California, Irvine, says that the field is still working to understand how the natural enzymes affected by NMN+ work, and researchers might need to optimize their bacteria on a case-by-case basis.
Beyond NMN
Other researchers are exploring different NRCs. Zongbao Kent Zhao, a bioengineer at the Dalian Institute of Chemical Physics in China, is engineering enzymes to use a cofactor called nicotinamide cytosine dinucleotide (NCD), which uses a cytosine nucleotide in place of the adenine in NAD. Unlike NMN, NCD is entirely artificial: cells do not naturally make it, although enzymes can be engineered to use it as a cofactor. Zhao hopes that this feature will prevent ‘crosstalk’ between the biosynthetic and natural metabolic pathways, which might be able to use NMN in place of NAD when levels of NMN are high, leaving less, therefore, for the desired synthesis reactions.
Zhao’s group has altered two of the enzymes in the E. coli pathway that produces malate4 — a precursor to lactic acid — so that they use NCD instead of NAD. His eventual goal, Zhao says, is to make biofuels such as biodiesel that can be “dropped in” to combustion engines without needing to modify the engines.
Decoupling natural from biosynthetic processes could make the manufacturing process more environmentally friendly, says Pablo Iván Nikel, a biotechnologist at the Technical University of Denmark in Kongens Lyngby. “This seems to be something the industry is craving.” For chemical manufacturers using microorganisms to spin natural substances into useful chemicals, it could solve two major problems, he says: the need for fossil fuels as substrates and the production of toxic waste by-products.
Nikel’s group, for instance, has been trying to find ways to get bacteria to produce fluorinated precursors of compounds such as polytetrafluoroethylene (Teflon), which are highly toxic to manufacture. His group focuses on Pseudomonas putida, a species that thrives in toxic conditions. In 2020, the group described genetic circuits that force this tough bacterium to add fluoride to sugars and other chemicals5 — a biological process that occurs rarely in nature. Now, the team is trying to incorporate NRCs into this process in the hope of making this difficult process more efficient, and eventually to drive reactions using carbon dioxide captured from the atmosphere. “We’re replacing this oil-dependent society with more ecofriendly production systems,” Nikel says.

Han Li is one of several researchers investigating non-canonical redox cofactors.Credit: Natalie Tso and Cynthia Dieudonne
NAD isn’t the only redox cofactor that could be replaced with synthetic substitutes. Stephan Hammer, an organic chemist at the University of Bielefeld in Germany, and his team are developing analogues for SAM, which participates in reactions that add methyl groups or sulfur groups called sulfonium to chemical compounds.
Sulfonium reactions are rare in nature, and Hammer says that being able to control them with synthetic SAM substitutes could enable researchers to produce pharmaceutical drugs or label proteins so that they can be tracked in cells. “If you potentially have new cofactors and new reactivities, you can expand on what’s in nature,” he says. But although his group has found ways to synthesize large amounts of different SAM analogues in cells, incorporating them into synthetic pathways has been difficult because the enzymes’ activity is still low, he says.
Science fiction … in cells
Indeed, NRCs present challenges on multiple levels, specialists say. Ideally, they should work with engineered enzymes but not interact with existing enzymes, says Caroline Paul, a biological chemist at the Delft University of Technology in the Netherlands. Her group is working on NMN with Weusthuis, engineering enzymes that use it exclusively rather than interchanging it with NAD. The researchers are also finding ways to modify NMN and other cofactors with chemical tails that tailor the compounds to certain enzymes such as monooxygenases, which cells use to create signalling molecules. Researchers could eventually exploit such designs to use different NRCs at different steps of the reaction in the same cell.
For now, Paul’s team is focusing on lactic acid and ethanol production because these systems are well characterized. From there, she hopes to move onto complex chemicals such as those used in perfumes.
And that’s where NRCs could be particularly useful, says Richard van Kranenburg, a biotechnologist at Amsterdam-based biochemicals company Corbion, the world’s leading producer of industrial lactic acid. “I don’t think we could improve lactic-acid production,” he says. Incorporating NRCs and engineered enzymes would require manufacturers to set up new production pathways to yield a chemical that microorganisms naturally make very efficiently.
But decreasing costs could make biomanufacturing more viable for chemicals such as the natural plant phytoestrogens that are used in some cancer drugs. For plants, phytoestrogen synthesis is a complex, multistep and inefficient process that involves many enzymes, says Jules Beekwilder, a chemical biologist with the chemical manufacturer BASF in Geleen, the Netherlands. Companies could put these enzymes into yeast or bacteria and perhaps produce the chemical in a fermentation tank, but the many production steps mean that energy is lost throughout the process. In those cases, increased output could justify the added expenses, van Kranenburg says. “The cost of your medium doesn’t matter and maybe even the cost of the cofactor does not matter if the product is valuable enough.”
Beekwilder and van Kranenburg are industry advisers for Weusthuis’s and Paul’s research teams, although neither BASF nor Corbion fund the research. Both advisers say that they are excited about the potential of NRCs. “It may be nice to control redox conditions without affecting the physiology of the organism,” Beekwilder says. But he adds that scaling them up to meet consumer demand presents its own set of challenges, such as maintaining the varying levels of oxygen needed for different enzymatic reactions. And although researchers often know what each enzyme does, they might not understand the biophysics well enough to be able to modify the enzyme to accept an NRC. “It’s not so that you can press the button on the computer and it’s done,” he says.
That said, advances in artificial intelligence (AI) are making enzyme engineering much easier than it was just a few years ago. “The idea that we have to crystallize a protein is ending,” Pfleger says, referring to the classic structural-biology technique of X-ray crystallography. “The ability to give proteins new functions or design from scratch is accelerating like crazy.” At the same time, AI tools such as AlphaFold and RoseTTAFold are learning how to decode enzymes’ structure from their sequence and could predict how specific modifications — a new fold that allows the enzyme to accept NMN instead of NAD, for instance — would affect its function.
NatureTech hub
Li says that she is “very excited” about the possibilities of AI and hopes one day to accumulate enough data to work out which modifications are needed to make any enzyme work with NRCs, and to start automating the process for creating them. Her group has already begun working out design principles that would allow enzymes to switch their preference from NAD to NMN.
Researchers might even be able to modify certain NRC-powered enzymes to be more powerful than their natural counterparts, or to work at different acidities, temperatures or oxygen levels from those that are normally required by natural enzymes. If the enzymes are versatile enough, Li estimates that NMN-powered reactions could end up being significantly cheaper than those that use NAD, owing to the increased efficiency and, potentially, the lower cost of producing NMN compared with NAD.
Still, researchers would prefer to be as efficient as possible. In nature, biosynthetic redox reactions are coupled with secondary reactions that recycle NADH to NAD+, or vice versa, so that the cell can keep the biosynthesis going. Research groups including Li’s are designing ‘recycling’ enzymes that can switch NMN+ to and from NMNH, creating oxidative or reductive conditions in the cell that are independent of NAD. Ultimately, Li hopes to find or develop a universal recycling enzyme or electrode that can switch any NRC back to its oxidized or reduced forms, depending on what the artificial biosynthetic pathway needs.
Not that every enzyme in a complex synthesis pathway would need to depend on an NRC, Hammer notes; the engineered enzymes could be reserved for particularly inefficient bottlenecks, or to make minor modifications to a compound that the cell has already made.
As the roster of NRCs expands, says Weusthuis, the biosynthetic possibilities are endless. “This is science fiction, not in space but in cells.”
[ad_2]
Source Article Link