[ad_1]
A nanopore sequencing device is typically used for sequencing DNA and RNA.Credit: Anthony Kwan/Bloomberg/Getty
With its fast analyses and ultra-long reads, nanopore sequencing has transformed genomics, transcriptomics and epigenomics. Now, thanks to advances in nanopore design and protein engineering, protein analysis using the technique might be catching up.
“All the pieces are there to start with to do single-molecule proteomics and identify proteins and their modifications using nanopores,” says chemical biologist Giovanni Maglia at the University of Groningen, the Netherlands. That’s not precisely sequencing, but it could help to work out which proteins are present. “There are many different ways you can identify proteins which doesn’t really require the exact identification of all 20 amino acids,” he says, referring to the usual number found in proteins.
In nanopore DNA sequencing, single-stranded DNA is driven through a protein pore by an electrical current. As a DNA residue traverses the pore, it disrupts the current to produce a characteristic signal that can be decoded into a sequence of DNA bases.
Proteins, however, are harder to crack. They cannot be consistently unfolded and moved by a voltage gradient because, unlike DNA, proteins don’t carry a uniform charge. They might also be adorned with post-translational modifications (PTMs) that alter the amino acids’ size and chemistry — and the signals that they produce. Still, researchers are making progress.
Water power
One way to push proteins through a pore is to make them hitch a ride on flowing water, like logs in a flume. Maglia and his team engineered a nanopore1 with charges positioned so that the pore could create an electro-osmotic flow that was strong enough to unfold a full-length protein and carry it through the pore. The team tested its design with a polypeptide containing negatively charged amino acids, including up to 19 in a row, says Maglia. This concentrated charge created a strong pull against the electric field, but the force of the moving water kept the protein moving in the right direction. “That was really amazing,” he says. “We really did not expect it would work so well.”
Super-speedy sequencing puts genomic diagnosis in the fast lane
Chemists Hagan Bayley and Yujia Qing at the University of Oxford, UK, and their colleagues have also exploited electro-osmotic force, this time to distinguish between PTMs2. The team synthesized a long polypeptide with a central modification site. Addition of any of three distinct PTMs to that site changed how much the current through the pore was altered relative to the unmodified residues. The change was also characteristic of the modifying group. Initially, “we’re going for polypeptide modifications, because we think that’s where the important biology lies”, explains Qing.
And, because nanopore sequencing leaves the peptide chain intact, researchers can use it to determine which PTMs coexist in the same molecule — a detail that can be difficult to establish using proteomics methods, such as ‘bottom up’ mass spectrometry, because proteins are cut into small fragments. Bayley and Qing have used their method to scan artificial polypeptides longer than 1,000 amino acids, identifying and localizing PTMs deep in the sequence. “I think mass spec is fantastic and provides a lot of amazing information that we didn’t have 10 or 20 years ago, but what we’d like to do is make an inventory of the modifications in individual polypeptide chains,” Bayley says — that is, identifying individual protein isoforms, or ‘proteoforms’.
Molecular ratchets
Another approach to nanopore protein analysis uses molecular motors to ratchet a polypeptide through the pore one residue at a time. This can be done by attaching a polypeptide to a leader strand of DNA and using a DNA helicase enzyme to pull the molecule through. But that limits how much of the protein the method can read, says synthetic biologist Jeff Nivala at the University of Washington, Seattle. “As soon as the DNA motor would hit the protein strand, it would fall off.”
Nivala developed a different technique, using an enzyme called ClpX (see ‘Read and repeat’). In the cell, ClpX unfolds proteins for degradation; in Nivala’s method, it pulls proteins back through the pore. The protein to be sequenced is modified at either end. A negatively charged sequence at one end allows the electric field to drive the protein through the pore until it encounters a stably folded ‘blocking’ domain that is too large to pass through. ClpX then grabs that folded end and pulls the protein in the other direction, at which point the sequence is read. “Much like you would pull a rope hand over hand, the enzyme has these little hooks and it’s just dragging the protein back up through the pore,” Nivala says.
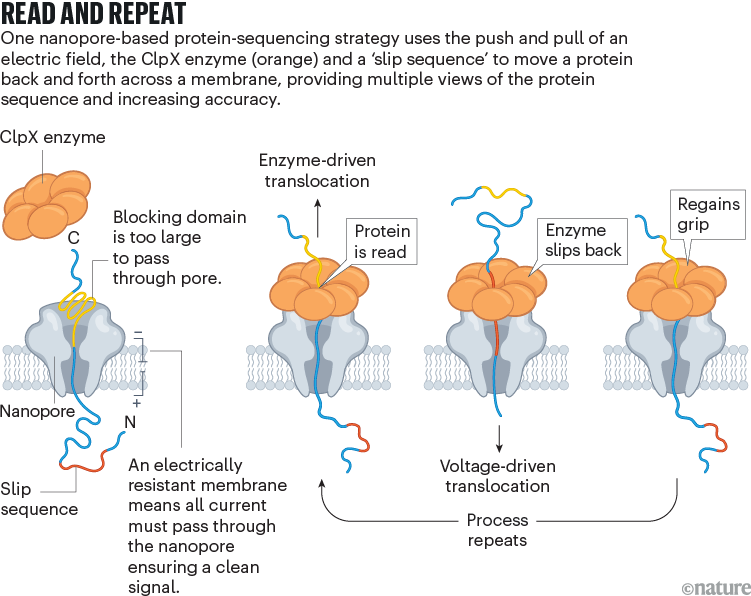
Source: Ref. 3
Nivala’s approach has another advantage: when ClpX reaches the end of the protein, a special ‘slip sequence’ causes it to let go so that the current can pull the protein through the pore for a second time. As ClpX reels it back out again and again, the system gets multiple peeks at the same sequence, improving accuracy.
Last October3, Nivala and his colleagues showed that their method can read synthetic protein strands of hundreds of amino acids in length, as well as an 89-amino-acid piece of the protein titin. The read data not only allowed them to distinguish between sequences, but also provided unambiguous identification of amino acids in some contexts. Still, it can be difficult to deduce the amino-acid sequence of a completely unknown protein, because an amino acid’s electrical signature varies on the basis of both its surrounding sequence and its modifications. Nivala predicts that the method will have a ‘fingerprinting’ application, in which an unknown protein is matched to a database of reference nanopore signals. “We just need more data to be able to feed these machine-learning algorithms to make them robust to many different sequences,” he says.
NatureTech hub
Stefan Howorka, a chemical biologist at University College London, says that nanopore protein sequencing could boost a range of disciplines. But the technology isn’t quite ready for prime time. “A couple of very promising proof-of-concept papers have been published. That’s wonderful, but it’s not the end.” The accuracy of reads needs to improve, he says, and better methods will be needed to handle larger PTMs, such as bulky carbohydrate groups, that can impede the peptide’s movement through the pore.
How easy it will be to extend the technology to the proteome level is also unclear, he says, given the vast number and wide dynamic range of proteins in the cell. But he is optimistic. “Progress in the field is moving extremely fast.”
[ad_2]
Source Article Link



