[ad_1]
Bacterial strains
Mtb strains are derivatives of H37Rv unless otherwise noted. ΔbioA Mtb was obtained from the Schnappinger laboratory64. E. coli strains are derivatives of DH5α (NEB), Rosetta2, or BL21(DE3) (Novagen).
Mycobacterial cultures
Mtb was grown at 37 °C in Difco Middlebrook 7H9 broth or on 7H10 agar supplemented with 0.2% glycerol (7H9) or 0.5% glycerol (7H10), 0.05% Tween-80, 1× oleic acid-albumin-dextrose-catalase (OADC) and the appropriate antibiotics (kanamycin 10–20 μg ml−1 and/or hygromycin 25–50 μg ml−1). ATc was used at 100 ng ml−1. Mtb cultures were grown standing in tissue culture flasks (unless otherwise indicated) with 5% CO2. Note that both 7H9 and 7H10 medium are normally supplemented with biotin (0.5 mg l−1; ~2 μM), thereby allowing growth of the ΔbioA Mtb auxotroph.
Selection of Rif-resistant Mtb isolates
For the selection of RifR H37Rv and ΔbioA Mtb, 5 independent 5-ml cultures were started at a density of ~2,000 cells per ml (to minimize the number of preexisting RifR bacteria) and grown to stationary phase (OD600 > 1.5). Cultures were pelleted at 4,000 rpm for 10 min, resuspended in 30 μl remaining medium per pellet and plated on 7H10 agar supplemented with Rif at 0.5 μg ml−1. After outgrowth, colonies were picked into 7H9 medium. After 1 week of outgrowth, an aliquot was heat-inactivated and the Rif resistance determining region of rpoB, rpoA and rpoC were amplified by PCR and Sanger sequenced. See Supplementary Table 4 for primer sequences.
Generation of structural models
The structural model of Mtb RNAP transcription initiation complex bound to Rif in Fig. 1a was generated by modelling Mycobacterium smegmatis RNAP bound to Rif (PDB: 6CCV)65 on to the transcription initiation complex structure (PDB: 6EDT)66.
The cryo-EM structures of a NusG-bound paused elongation complex from Mtb (PDB: 8E74) in Fig. 2d, and the location of clinical isolate mutations in Fig. 4a are derived from Delbeau et al.13.
Generation of individual CRISPRi strains
Individual CRISPRi plasmids were cloned as described67 using Addgene plasmid 166886. In brief, the CRISPRi plasmid backbone was digested with BsmBI-v2 (NEB R0739L) and gel-purified. sgRNAs were designed to target the non-template strand of the target gene open reading frame (ORF). For each individual sgRNA, two complementary oligonucleotides with appropriate sticky end overhangs were annealed and ligated (T4 ligase NEB M0202 M) into the BsmBI-digested plasmid backbone. Successful cloning was confirmed by Sanger sequencing.
Individual CRISPRi plasmids were then electroporated into Mtb. Electrocompetent cells were obtained as described68. In brief, an Mtb culture was expanded to an OD600 = 0.4–0.6 and treated with glycine (final concentration 0.2M) for 24 h before pelleting (4,000g for 10 min). The cell pellet was washed three times in sterile 10% glycerol. The washed bacilli were then resuspended in 10% glycerol in a final volume of 5% of the original culture volume. For each transformation, 100 ng plasmid DNA and 100 μl electrocompetent mycobacteria were mixed and transferred to a 2 mm electroporation cuvette (Bio-Rad 1652082). Where necessary, 100 ng plasmid plRL19 (Addgene plasmid 163634) was also added. Electroporation was performed using the Gene Pulser X cell electroporation system (Bio-Rad 1652660) set at 2,500 V, 700 Ω and 25 μF. Bacteria were recovered in 7H9 for 24 h. After the recovery incubation, cells were plated on 7H10 agar supplemented with the appropriate antibiotic to select for transformants.
CRISPRi library transformation
CRISPRi libraries were generated as described previously28. In brief, fifty transformations were performed to generate RifS and βS450L ΔbioA libraries. For each transformation, 1 μg of RLC12 plasmid DNA was added to 100 μl electrocompetent cells. The cells:DNA mix was transferred to a 2 mm electroporation cuvette (Bio-Rad 1652082) and electroporated at 2,500 kV, 700 Ω, and 25 μF. Each transformation was recovered in 2 ml 7H9 medium supplemented with OADC, glycerol and Tween-80 (100 ml total) for 16–24 h. The recovered cells were collected at 4,000 rpm for 10 min, resuspended in 400 μl remaining medium per transformation and plated on 7H10 agar supplemented with kanamycin (see ‘Mycobacterial cultures’) in Corning Bioassay dishes (Sigma CLS431111-16EA).
After 21 days of outgrowth on plates, transformants were scraped and pooled. Scraped cells were homogenized by two dissociation cycles on a gentleMACS Octo Dissociator (Miltenyi Biotec 130095937) using the RNA_01 program and 30 gentleMACS M tubes (Miltenyi Biotec 130093236). The library was further declumped by passaging 1 ml of homogenized library into 100 ml of 7H9 supplemented with kanamycin (see Mycobacterial cultures) for between 5 and 10 generations. Final RifS and βS450L ΔbioA Mtb library stocks were obtained after passing the cultures through a 10-μm cell strainer (Pluriselect SKU 43-50010-03). Genomic DNA was extracted from the final stocks and library quality was validated by deep sequencing (see ‘Genomic DNA extraction and library preparation for Illumina sequencing’).
Pooled CRISPRi screen
Pooled CRISPRi screens were performed as described28. In brief, 20-ml cultures were grown in vented tissue culture flasks (T-75; Falcon 353136) and 7H9 medium supplemented with kanamycin (see ‘Mycobacterial cultures’) and maintained at 37 °C, 5% CO2 in a humidified incubator.
The screen was initiated by thawing 4× 1-ml aliquots of the Mtb ΔbioA (RifS or βS450L) CRISPRi library (RLC12) and inoculating each aliquot into 24 ml 7H9 medium supplemented with kanamycin in a T-75 flask (starting OD600∼0.06). The cultures were expanded to approximately OD600 = 1.0, pooled and passed through a 10-μm cell strainer (pluriSelect 43-50010-03) to obtain a single cell suspension. The single cell suspension (flow-though) was used to set up six ‘generation 0’ cultures: three replicate cultures with ATc (+ATc) and three replicate control cultures without ATc (–ATc). From each generation 0 culture, we collected 10 OD600 units of bacteria (∼3 × 109 bacteria; ∼30,000X coverage of the CRISPRi library) for genomic DNA extraction. The remaining culture volume was used to initiate the pooled CRISPRi fitness screen. Cultures were periodically passaged in pre-warmed medium in order to maintain log phase growth. At generation 2.5, 5, and 7.5, cultures were back-diluted 1:6 (to a starting OD600 = 0.2) and cultivated for approximately 2.5 doublings. At generation 10, 15, 20, and 25, cultures were back-diluted 1:24 (to a starting OD600 = 0.05) and expanded for 5 generations before reaching late-log phase. ATc was replenished at every passage. By keeping the OD600 of the 20 ml cultures ≥ 0.05, we guaranteed sufficient coverage of the library (3,000X) at all times. At set time points (approximately 2.5; 5; 7.5; 10; 15; 20; 25 and 30 generations), we collected bacterial pellets (10 OD600 units) to extract genomic DNA.
Genomic DNA extraction and library preparation for Illumina sequencing of CRISPRi libraries
Genomic DNA was isolated from bacterial pellets using the CTAB-lysozyme method described previously69. Genomic DNA concentration was quantified using the DeNovix dsDNA high sensitivity assay (KIT-DSDNA-HIGH-2; DS-11 Series Spectrophotometer/Fluorometer).
Illumina libraries were constructed as described28. In brief, the sgRNA-encoding region was amplified from 500 ng genomic DNA using NEBNext Ultra II Q5 master Mix (NEB M0544L). PCR cycling conditions were: 98 °C for 45 s; 17 cycles of 98 °C for 10 s, 64 °C for 30 s, 65 °C for 20 s; 65 °C for 5 min. Each PCR reaction a unique indexed forward primer (0.5 μM final concentration) and a unique indexed reverse primer (0.5 μM) (Supplementary Table 4). Forward primers contain a P5 flow cell attachment sequence, a standard Read1 Illumina sequencing primer binding site, custom stagger sequences to ensure base diversity during Illumina sequencing, and unique barcodes to allow for sample pooling during deep sequencing. Reverse primers contain a P7 flow cell attachment sequence, a standard Read2 Illumina sequencing primer binding site, and unique barcodes.
Following PCR amplification, each ∼230 bp amplicon was purified using AMPure XP beads (Beckman–Coulter A63882) using two-sided selection (0.75X and 0.12X). Eluted amplicons were quantified with a Qubit 2.0 Fluorometer (Invitrogen), and amplicon size and purity were quality controlled by visualization on an Agilent 4200 TapeStation (Instrument- Agilent Technologies G2991AA; reagents- Agilent Technologies 5067-5583; tape- Agilent Technologies 5067-5582). Next, individual PCR amplicons were multiplexed into 20 nM pools and sequenced on an Illumina sequencer according to the manufacturer’s instructions. To increase sequencing diversity, a PhiX spike-in of 2.5–5% was added to the pools (PhiX sequencing control v3; Illumina FC-110-3001). Samples were run on the Illumina NextSeq 500 or NovaSeq 6000 platform (single-read 1 ×85 cycles, 8 × i5 index cycles, and 8 × i7 index cycles).
Differential vulnerability analysis of Rif-resistant versus Rif-sensitive strains
Gene vulnerability in the RifS and βS450L Mtb strains was determined using an updated vulnerability model based on the one previously described28. In the updated model, read counts for a given sgRNA in the minus ATc conditions were modelled using a negative binomial distribution with a mean proportional to the counts in the plus ATc condition, plus a factor representing the log2 fold change:
$${y}_{i}^{-{\rm{ATc}}} \sim {\rm{NegBinom}}\left({\eta }_{i},\phi \right)$$
$${\eta }_{i}=\log (\,{y}_{i}^{+{\rm{ATc}}}+{\lambda }_{i})+{\rm{TwoLine}}({x}_{i},{\alpha }_{l},{\beta }_{l},\gamma ,{\beta }_{e})$$
where λi is an sgRNA-level correction factor estimated by the model, xi represents the generations analysed for the ith guide, and the TwoLine function represents the piecewise linear function previously described, which models sgRNA behaviour over the logistic function describing gene-level vulnerabilities was simplified by setting the top asymptote of the curve (previously K) equal to 0, representing the fact that weakest possible sgRNAs are expected to impose no effect on bacterial fitness, that is:
$${\rm{Logistic}}\left(s\right)=\frac{{\beta }_{\max }}{\left(1+{{\rm{e}}}^{\left(-H\cdot \left(s-M\right)\right)}\right)}$$
The Bayesian vulnerability model was run for each condition independently, and samples for all the parameters were obtained using Stan running 4 independent chains with 1,000 warmup iterations and 3,000 samples each (for a total of 12,000 posterior samples for each parameter in the model after discarding warmup iterations).
Differential vulnerabilities were estimated by two approaches. First, for each gene, the difference in pairwise (guide-level) vulnerability estimates was obtained, resulting in posterior samples of the differential vulnerability (delta-vulnerability). This effectively estimated the difference in the integrals of the vulnerability functions. If the 95% credible region did not overlap 0.0 those were taken as significant differential vulnerabilities between the strains.
Next, to identify differences between genes which may not exhibit the expected dose–response curve, we estimated the fitness cost (log2FC) predicted by our model for a (theoretical) sgRNA of strength 0.0 (that is, Logistic(s = 0)). This represented the weakest phenotype theoretically possible with our CRISPRi system, which we call Fmin. The difference between this value was estimated for each gene (∆Fmin) and those where the 95% credible region did not overlap 0.0 were identified as significant differential vulnerabilities by this approach.
Pathway analysis
First, all annotated Mtb genes were associated with a pathway as defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database70,71,72. If necessary, annotations were manually curated to update or correct pathway assignments. To quantify pathway enrichment, the query set was defined as the union of the upper quartile of differential vulnerabilities defined by both the original gene vulnerability calling method (ΔV) and the Fmin approach. The background set was defined as all annotated Mtb genes. Enrichment of the pathways identified as differentially vulnerable was calculated by an odds ratio and significance was determined with a Fisher’s exact test.
phyOverlap
To detect associations between gene variants and Rif resistance, we employed a phylogenetic convergence test using the phyOverlap algorithm73 (https://github.com/Nathan-d-hicks/phyOverlap). In brief, FASTQ files were aligned to H37Rv genome (NC_018143.2) using bwa (version 0.7.17-r1188). FASTQ accession numbers are provided in Supplementary Table 3. Single-nucleotide polymorphisms (SNPs) were called and annotated using the HaplotypeCaller tool Genome Analysis Toolkit (version 3.5) using inputs from samtools (version 1.7). SNP sites with less than 10x coverage or missing data in >10% strains were removed from the analysis. Repetitive regions of the genome (PE/PPE genes, transposases, and prophage genes) are excluded from the analysis. Known drug-resistance regions were further excluded so as not to bias phylogenetic tree construction. M. canetti was provided as an outgroup (NC_015848). We performed Maximum Likelihood Inference using RAxML (v8.2.11) to construct the ancestral sequence and determine the derived state of each allele. Overlap with Rif resistance was scored by dividing the number of genotypically predicted (Mykrobe v0.9.012) RifR isolates containing a derived allele by the total number of isolates with a derived allele at a given genomic position. To generate a gene-wide score, we excluded synonymous SNPs and averaged the individual nonsynonymous SNP scores, weighting the scores by the number of times derived alleles evolved across the phylogenetic tree. The significance of the overlap is then tested by redistributing mutation events for each SNP randomly across the tree and recalculating the score. This permutation is done 50,000 times to derive the P value. This analysis additionally used FastTree (version 2.1.11) and figTree (v1.4.4).
dN/dS calculations
The ratio of nonsynonymous (dN) to synonymous (dS) nucleotide substitutions was used to quantify selective pressure acting on nusG and rpoC. A dN/dS value less than one suggests negative or purifying selection whereas a dN/dS value greater than one suggests positive or diversifying selection. For this analysis, we used a collection of ~50,000 Mtb clinical isolate whole-genome sequences, as described41. Isolates were grouped based on the presence of genotypically predicted Rif resistance (Mykrobe v0.9.012), as well as the identity of the rpoB mutation (S450X or H445X; where X indicates any amino acid other than Ser or His, respectively) conferring RifR. The number of samples used in the nusG dN/dS analysis shown in Fig. 3 are as follows: 1,365 RifS, 350 RifR, 270 S450X, and 26 H445X. The number of samples used in the rpoC dN/dS analysis shown in Fig. 3 are as follows: 23,024 RifS, 13,993 RifR, 11,067 S450X, and 1,215 H445X. Insertions and deletions were necessarily excluded from this analysis. A bootstrap-analysis was performed to calculate the dN/dS ratios to reduce any potential effects of recent clonal expansion events or convergent evolution of a specific site, like acquired drug-resistance mutations, as performed previously44. The analysis was performed by sub-sampling 80% of total variants in each group. The sub-sampling was repeated 100 times. dN/dS values were calculated for each subset of samples using a python script obtained from the github repository: https://github.com/MtbEvolution/resR_Project/tree/main/dNdS.
SNP calling and upset plot
SNP information for all Mtb clinical isolate whole-genome sequences were called as follows. FASTQ reads were aligned to the H37Rv genome (NC_018143.2) and SNPs were called and annotated using Snippy9 (version 3.2-dev) using default parameters (minimum mapping quality of 60 in BWA, samtools base quality threshold of 20, minimum coverage of 10, minimum proportion of reads that differ from reference of 0.9). Mapping quality and coverage was further assessed using QualiMap with the default parameters (version 2.2.2-dev). Samples with a mean coverage < 30, mean mapping quality ≤ 45, or GC content ≤ 50% or ≥ 70% were excluded. Drug resistance-conferring SNPs were annotated using Mykrobe (v0.9.012). The resulting SNP and drug-resistance calls were used to generate the values depicted in the upset plot.
Phylogenetic trees
Phylogenetic trees based on SNP calls described above were built using FastTree (version 2.1.11 SSE3). A list of SNPs in essential genes was concatenated to build phylogenetic trees. Indels, drug resistance-conferring SNPs, and SNPs in repetitive regions of the genome (PE/PPE genes, transposases and prophage genes) were excluded. Tree visualization was performed in iTol (https://itol.embl.de/).
Barcode library production
The barcode library was designed to include over 100,000 random 18-mer sequences cloned into an Giles-integrating backbone (attP only, no Integrase) containing a hygromycin resistance cassette with a premature stop codon (plNP472). Oligonucleotides were synthesized as a gBlocks Library by IDT, containing 104,976 fragments.
plNP472 (1.6 μg) was digested with PciI (NEB R0655) and gel-purified (QIAGEN 28706). The library was PCR amplified using NEBNext High-Fidelity 2X PCR Master Mix (NEB M0541L). One 50-μl reaction was prepared, containing 25 μl of PCR master mix, 0.0125 pmol of the gBlock library, and a final concentration of 0.5 μM of the appropriate forward and reverse primers (Fwd: 5′-TTACGCGTTTCACTGGCCGATTG-3′ + Rev: 5′-TTTTGCTGGCCTTTTGCTCAAC-3′). PCR cycling conditions were: 98 °C for 30 s; 15 cycles of 98 °C for 10 s, 68 °C for 10 s, 72 °C for 15 s; 72 °C for 120 s. The PCR amplicon were purified using the QIAGEN MinElute PCR purification kit (QIAGEN 28004). One Gibson assembly reaction (NEB E2621) was prepared with 0.01 pmol μl−1 digested plNP472 backbone, 0.009 pmol μl−1 cleaned PCR amplicon, and master mix, representing a 1:2 molar ratio of vector:insert.
Following incubation at 50 °C for 1 h, 7 μl the Gibson product was dialysed to remove salts and transformed into 100 μl MegaX DH10B T1R Electrocomp Cells (Invitrogen C640003) diluted with 107 μl 10% glyerol. For each of three total transformations, 75 μl of the cells:DNA mix was transferred to a 0.1 cm electroporation cuvette (Bio-Rad 1652089) and electroporated at 2,000 V, 200 ohms, 25 μF. Transformations were washed twice with 300 μl provided recovery medium and recovered in a total of 3 ml medium. Cells were allowed to recover at 37 °C with gentle rotation. Recovered cells were plated across three plates of LB agar supplemented with zeocin. After 1 d incubation at 37 °C, transformants were scraped and pooled. One fourth of the pellet (3.2 g dry mass) was used to perform 24 minipreps using a QIA prep Spin Miniprep Kit (Qiagen 27104).
Transformation of barcode library into Mtb
The barcode library was transformed into RifS and βS450L Mtb expressing RecT (mycobacteriophage recombinase) similarly to the CRISPRi library (see CRISPRi library transformation), with minor modifications. In brief, cultures for competent cells were grown in 7H9 supplemented with kanamycin to retain the episomal recT encoding plasmid (plRL4). Twenty-millilitre cultures were concentrated ten times and transformed with 250 ng of library and 100 ng of non-replicating, Giles integrase containing plasmid (plRL40). Additionally, after recovery cells were plated on 7H10 agar supplemented with kanamycin and zeocin. Transformants were scrapped after 29 days of outgrowth.
ssDNA recombineering and validation of strains
Clinical nusG, rpoB and rpoC mutants were introduced into RifS and βS450L Mtb using oligonucleotide-mediated (ssDNA) recombineering, as described previously68. In brief, 70-mer oligonucleotides were designed to correspond to the lagging strand of the replication fork, with the desired mutation in the middle of the sequence. Alterations were chosen to avoid recognition by the mismatch-repair machinery of RecT expression was induced ~16 h before transformation by addition of ATc to a final concentration of 0.5 μg ml−1. 400 μl of competent cells were transformed with 5 μg of mutation containing oligonucleotide and 0.1 μg of hygromycin resistance cassette repair oligonucleotide (1:50 ratio of mutant oligonucleotide to repair oligonucleotide) and recovered in 5 ml 7H9 medium.
After 24 h of recovery, 200 μl of cells were plated on 7H10 plates supplemented with hygromycin. After 21 days of outgrowth, 12 colonies per construct were picked into 100 μl 7H9 medium supplemented with hygromycin in a 96 well plate (Fischer Scientific 877217). 50 μl of culture were heat-inactivated at 80 °C for 2 h in a sealed microamp 96 well plate (Fischer Scientific 07200684; Applied Biosystems N8010560). Fifty microlitres of heat-inactivated culture was mixed with 50 μl of 25% DMSO and lysed at 98 °C 10 min.
Mutations of interest and unique barcodes were confirmed with PCR amplification and Sanger sequencing. The region of interest was PCR amplified with NEBNext High-Fidelity 2X PCR Master Mix (NEB M0541L) using 0.5 μl of heat-lysed product with the appropriate primers, annealing temperatures and extension times (see Supplementary Table 4). Residual PCR primers were removed with NEB Shrimp Alkaline Phosphatase (rSAP) and exonuclease I (exo) (rSAP- NEB M0371; exo- NEB M0293) per manufacturer’s instructions. Amplicons were then submitted for Sanger sequencing. One to three unique independent isolates were generated for all tested mutations.
Pooled barcode competitive growth assay
Validated mutants were first grown in 1 ml 7H9 with hygromycin and after 3 days, expanded to 5 ml 7H9 with hygromycin. Strains were pooled to contain approximately 1.2 × 107 cells for each mutant. The pool was then diluted to a starting OD600 of 0.01 in 7H9 supplemented with hygromycin. At this point, three 20 ml cultures in vented tissue culture flasks (T-75; Falcon 353136) were expanded to late-log phase and used as input for the competitive growth experiment. Sixteen OD600 units of cells were collected from flask as the input culture (generation 0). Triplicate cultures were then diluted back to OD600 = 0.05 and grown for ~4.5 generations, back-diluted again to OD600 = 0.05 and grown for an additional 4 generations. After this, cultures were collected for a cumulative 8.5 generations of competitive growth.
Genomic DNA extraction and library preparation for next-generation sequencing followed the same protocol as that of the CRISPRi libraries (see above), with minor modifications. In brief, the barcode region was amplified from 100 ng genomic DNA using NEBNext Ultra II Q5 master Mix (NEB M0544L). PCR cycling conditions were: 98 °C for 45 s; 16 cycles of 98 °C for 10 s, 64 °C for 30 s, 65 °C for 20 s; 65 °C for 5 min. Each PCR reaction contained a unique indexed forward primer (0.5 μM final concentration) and a unique indexed reverse primer (0.5 μM) (see Supplementary Table 4). Additionally, individual PCR amplicons were multiplexed into a 1 nM pool and sequenced on an Illumina sequencer according to the manufacturer’s instructions. To increase sequencing diversity, a PhiX spike-in of 20% was added to the pool (PhiX sequencing control v3; Illumina FC-110-3001). Samples were run on the Illumina MiSeq Nano platform (paired-read 2 ×150 cycles, 8 × i5 index cycles, and 8 × i7 index cycles).
WGS and SNP calling for passaging timepoints and ssDNA recombinants
Genomic DNA (gDNA) was extracted as described above. gDNA was diluted and subjected to Illumina whole-genome sequencing by SeqCenter. In brief, Illumina libraries were generated through tagmentation-based and PCR-based Illumina DNA Prep kit and custom IDT 10 bp unique dial indices, generating 320 bp amplicons. Resulting libraries were sequenced on the Illumina NovaSeq 6000 platform (2 × 150 cycles). Demultiplexing quality control, and adapter trimming was performed with bcl-convert (v4.1.5).
Reads were aligned to the Mtb (H37Rv; CP003248.2) reference genome using bwa (v1.3.1) with default parameters. Variant detection was performed by Snippy (v4.6.0)/freebayes (v1.3.1). Resulting vcf files were inspected for compensatory mutations (Supplementary Table 2) in rpoABC and/or the presence of the desired mutation.
Definition of putative compensatory nusG, rpoA, rpoB, rpoC variants
Compensatory mutations in rpoA, rpoB and rpoC were taken from published sources and are described in Supplementary Table 2. Inclusion as a putative compensatory mutation in our list required that each reported variant in rpoA, rpoB, or rpoC was found specifically in Rif-resistant strains, defined here as meaning that ≥90% of all strains harbouring the putative compensatory mutation were genotypically predicted (gDST) RifR. The use of the ≥90% gDST RifR cut-off allows for presumptive instances of incorrect gDST calls for strains harbouring rare compensatory variants. The strains used for this analysis are the approximately 50,000 Mtb WGS strain collection described previously41.
The rules to define putative compensatory nusG mutations are as follows. Each nusG variant observed was assessed according to the following three rules and, if it met one of them, was deemed a putative compensatory variant.
-
(1)
The nusG variant was found in ≥80% genotypically predicted (gDST) RifR strains and was present in at least two distinct Mtb (sub)lineages. The use of the ≥80% gDST RifR cut-off allows for presumptive instances of incorrect gDST calls for strains harbouring rare nusG variants.
-
(2)
The nusG variant was found in 100% gDST RifR strains but only present in a single Mtb sublineage, but the same or nearby NusG site (±5 amino acids) was also mutated to an alternative amino acid that met the criteria stated in rule 1.
-
(3)
Residues based on the Mtb NusG–RNAP structure13 that were predicted to be important for the NusG pro-pausing activity (for example, NusG Trp120).
The rules to define a putative compensatory mutation in the rpoB β-protrusion were similar to those described for nusG, except that only rpoB β-protrusion residues at or near the NusG interface (RpoB Arg392–Thr410) were included in the analysis. Note that two such β-protrusion mutations (Thr400Ala and Gln409Arg) were previously identified as putative compensatory mutation17,74,75 (Supplementary Table 2).
RifR rpoB allele frequency distribution calculations
To check whether the observed distribution of RifR rpoB mutations was different for each of the three groups (all RifR strains in our clinical strain genome database, those harbouring known compensatory mutations in rpoA or rpoC, or those harbouring compensatory mutations in nusG or the β-protrusion), we performed a chi-squared test on the observed RifR rpoB mutant frequencies. Specifically, we take the RifR rpoB mutant frequencies observed in all RifR samples as representing an estimate of the base probabilities under the null hypothesis. We then use these base probabilities to calculate the frequency of mutations that would be expected in the other groups, based on the null hypothesis. That is:
For each mutation (m):
$$p(m)=\frac{{\rm{Number}}\,{\rm{of}}\,{\rm{times}}\,m\,{\rm{occurs}}\,{\rm{in}}\,{\rm{RifR}}\,{\rm{samples}}}{{\rm{Total}}\,{\rm{number}}\,{\rm{of}}\,{\rm{RifR}}\,{\rm{samples}}}$$
For each group (G) and mutation (m),
$$E\left[m| G\right]=p\left(m\right)\times {\rm{total}}\,{\rm{number}}\,{\rm{of}}\,{\rm{samples}}\,{\rm{in}}\,G$$
Protein expression and purification
Mtb RNAP
Mtb RNAP was purified as previously described66,76. In brief, plasmid pMP61 (wild-type RNAP) or pMP62 (S450L RNAP) was used to overexpress Mtb core RNAP subunits rpoA, rpoZ, a linked rpoBC and a His8 tag. pMP61/pMP62 was grown in E. coli Rosetta2 cells in LB with 50 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol at 37 °C to an OD600 of 0.3, transferred to room temperature and left shaking to an approximate OD600 of 0.6. RNAP expression was induced by adding IPTG to a final concentration of 0.1 mM, grown for 16 h, and collected by centrifugation (8,000g, 15 min at 4 °C). Collected cells were resuspended in 50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1 mM PMSF, 1 mM protease inhibitor cocktail, 5% glycerol and lysed by sonication. The lysate was centrifuged (27,000g, 15 min, 4 °C) and polyethyleneimine (PEI, Sigma-Aldrich) added to the supernatant to a final concentration of 0.6% (w/v) and stirred for 10 min to precipitate DNA binding proteins including target RNAP. After centrifugation (11,000g, 15 min, 4 °C), the pellet was resuspended in PEI wash buffer (10 mM Tris-HCl, pH 7.9, 5% v/v glycerol, 0.1 mM EDTA, 5 mM DTT, 300 mM NaCl) to remove non-target proteins. The mixture was centrifuged (11,000g, 15 min, 4 °C), supernatant discarded, then RNAP eluted from the pellet into PEI Elution Buffer (10 mM Tris-HCl, pH 7.9, 5% v/v glycerol, 0.1 mM EDTA, 5 mM DTT, 1 M NaCl). After centrifugation, RNAP was precipitated from the supernatant by adding (NH4)2SO4 to a final concentration of 0.35 g l−1. The pellet was dissolved in Nickel buffer A (20 mM Tris pH 8.0, 5% glycerol, 1 M NaCl, 10 mM imidazole) and loaded onto a HisTrap FF 5 ml column (GE Healthcare Life Sciences). The column was washed with Nickel buffer A and then RNAP was eluted with Nickel elution buffer (20 mM Tris, pH 8.0, 5% glycerol, 1 M NaCl, 250 mM imidazole). Eluted RNAP was subsequently purified by gel filtration chromatography on a HiLoad Superdex 26/600 200 pg in 10 mM Tris pH 8.0, 5% glycerol, 0.1 mM EDTA, 500 mM NaCl, 5 mM DTT. Eluted samples were aliquoted, flash frozen in liquid nitrogen and stored in −80 °C until usage.
Mtb σA–RbpA
Mtb σA–RbpA was purified as previously described76,77. The Mtb σA expression vector pAC2 contains the T7 promoter, ten histidine residues, and a precision protease cleavage site upstream of Mtb σA. The Mtb RbpA vector is derived from the pET-20B backbone (Novagen) and contains the T7 promoter upstream of untagged Mtb RbpA. Both plasmids were co-transformed into E. coli Rosetta2 cells and selected on medium containing kanamycin (50 µg ml−1), chloramphenicol (34 µg ml−1) and ampicillin (100 µg ml−1). Protein expression was induced at OD600 of 0.6 by adding IPTG to a final concentration of 0.5 mM and leaving cells to grow at 30 °C for 4 h. Cells were then collected by centrifugation (4,000g, 20 min at 4 °C). Collected cells were resuspended in 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole, 0.1 mM PMSF, 1 mM protease inhibitor cocktail, and 1 mM β-mercaptoethanol, then lysed using a continuous-flow French press. The lysate was centrifuged twice (15,000g, 30 min, 4 °C) and the proteins were purified by Ni2+-affinity chromatography (HisTrap IMAC HP, GE Healthcare Life Sciences) via elution at 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 500 mM imidazole, and 1 mM β-mercaptoethanol. Following elution, the complex was dialysed overnight into 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole, and 1 mM β-mercaptoethanol and the His10 tag was cleaved with precision protease overnight at a ratio of 1:30 (protease mass:cleavage target mass). The cleaved complex was loaded onto a second Ni2+-affinity column and was retrieved from the flow-through. The complex was loaded directly onto a size-exclusion column (SuperDex-200 16/16, GE Healthcare Life Sciences) equilibrated with 50 mM Tris-HCl, pH 8, 500 mM NaCl, and 1 mM DTT. The sample was concentrated to 4 mg ml−1 by centrifugal filtration and stored at –80 °C until usage.
Mtb CarD
Mtb CarD was purified as previously described66,76. In brief, Mtb CarD was overexpressed from pET SUMO (Invitrogen) in E. coli BL21(DE3) cells (Novagen) and selected on medium containing 50 µg ml−1 kanamycin. Protein expression was induced by adding IPTG to a final concentration of 1 mM when cells reached an apparent OD600 of 0.6, followed by 4 h of growth at 28 °C, then collected by centrifugation (4,000g, 15 min at 4 °C). Collected cells were resuspended in 20 mM Tris-HCl, pH 8.0, 150 mM potassium glutamate, 5 mM MgCl2, 0.1 mM PMSF, 1 mM protease inhibitor cocktail, and 1 mM β-mercaptoethanol, then lysed using a continuous-flow French press. The lysate was centrifuged twice (16,000g, 30 min, 4 °C) and the proteins were purified by Ni2+-affinity chromatography (HisTrap IMAC HP, GE Healthcare Life Sciences) via elution at 20 mM Tris-HCl, pH 8.0, 150 mM potassium glutamate, 250 mM imidazole, and 1 mM β-mercaptoethanol. Following elution, the complex was dialysed overnight into 20 mM Tris-HCl, pH 8.0, 150 mM potassium glutamate, 5 mM MgCl2, and 1 mM β-mercaptoethanol and the His10 tag was cleaved with ULP-1 protease (Invitrogen) overnight at a ratio of 1/30 (protease mass/cleavage target mass). The cleaved complex was loaded onto a second Ni2+-affinity column and was retrieved from the flow-through. The complex was loaded directly onto a size-exclusion column (SuperDex-200 16/16, GE Healthcare Life Sciences) equilibrated with 20 mM Tris-HCl, pH 8, 150 mM potassium glutamate, 5 mM MgCl2 and 2.5 mM DTT. The sample was concentrated to 5 mg ml−1 by centrifugal filtration and stored at –80 °C.
Wild-type Mtb NusG (+ mutants N65H, R124L and N125S)
Plasmid pAC82 (or mutant variation) was used to overexpress wild-type Mtb NusG13. Plasmids encoding NusG mutants were generated using Q5 Site-directed mutagenesis (NEB) and sequenced to confirm the presence of target mutations. E. coli BL21 cells containing plasmids encoding different versions of Mtb NusG were grown in LB with 50 μg ml−1 kanamycin at 37 °C to an OD600 of 0.4, then transferred to room temperature and left shaking to an OD600 of 0.67. Protein expression was induced by adding IPTG to a final concentration of 0.1 mM, grown for an additional 4 h, then collected by centrifugation (4,000g, 20 min at 4 °C). Collected cells were resuspended in 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole, 10% glycerol, 1 mM PMSF, 1 mM protease inhibitor cocktail (Roche), 2 mM β-mercaptoethanol, and lysed by French press. The lysate was centrifuged (4,000 rpm for 20 min, 4 °C) and the supernatant was removed and applied to a HisTrap column pre-washed with 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 15 mM imidazole, and 2 mM β-mercaptoethanol. After loading the sample, the column was washed with five volumes of the same buffer, before gradient elution with 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 250 mM imidazole, and 2 mM β-mercaptoethanol. The eluted protein was mixed with precision protease and dialysed overnight at 4 °C in 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10 mM β-mercaptoethanol to cleave the N-terminal His10 tag before applying to a HisTrap column to remove the uncleaved protein. The flow-through was collected and glycerol was added to a final concentration of 20% (v/v). Aliquots were flash frozen in liquid nitrogen and stored in –80 °C until use.
Promoter-based in vitro termination assays
The DNA sequence for the Mtb H37Rv 5 S rRNA (rrf gene) intrinsic terminator was taken from Mycobrowser (MTB000021), with genomic coordinates of 1,476,999 to 1,477,077 basepairs. The intrinsic terminator was found by predicting its RNA structure using mfold (RNA folding form v2.3) via the UNAFold Web Server. The intrinsic terminator was cloned downstream of a cytidine-less halt cassette in plasmid pAC7038, a gift of the R. Landick laboratory, using Q5 site-directed mutagenesis (following manufacturer’s protocol – NEB) at an annealing temperature of 59 °C with GC enhancer for the PCR step, with primers 5′-TGGTGTTTTTGTATGTTTATATCGACTCAGCCGCTCGCGCCATGGACGCTCTCCTGA-3′ and 5′-CCGTTACCGGGGGTGTTTTTGTATGTTCGGCGGTGTCCTGGATCCTGGCAGTTCCCT-3′ (synthesized by IDT), to create plasmid pJC1. The 323 base pairs linear DNA fragment used for in vitro transcription assays was PCR amplified using Accuprime Pfx DNA polymerase (Invitrogen) at an annealing temperature of 56.5 °C, with primers 5′-GAATTCAAATATTTGTTGTTAACTCTTGACAAAAGTGTTAAAAGC-3′ and 5′-GTTGCTTCGCAACGTTCAAATCC-3′ (synthesized by IDT), following manufacturer instructions, and PCR purified (using the QIAquick PCR Purification Kit (QIAGEN)) to remove protein contents and buffer exchange into 10 mM Tris-HCl pH 8.5.
pJC1 contains the rrf termination site at approximately +150 bp. This template also contained a C-less cassette (+1 to +26). Core RNAP was incubated for 15 min at 37 °C with σA/RbpA in transcription buffer (20 mM Tris, 25 mM KGlu, 10 mM MgOAc, 1 mM DTT, 5 µg ml−1 BSA) to form holo-RNAP, followed by 10 min incubation with 500 nM CarD at 37 °C. Holo-RNAP (200 nM) was then incubated with template DNA (10 nM) for 15 min at 37 °C. To initiate transcription, the complex was incubated with ATP + GTP (both at 16 µM), UTP (2 µM), and 0.1 µl per reaction [α-32P]UTP for 15 min at 37 °C to form a halted complex at U26. Transcription was restarted by adding a master mix containing NTP mix (A + C + G + U), heparin, and NusG at a final concentration of 150 µM (each NTP), 10 µg ml−1 (heparin), and 1 µM NusG at 23 °C. The reaction was allowed to proceed for 30 min, followed by a ‘chase’ reaction in which all 4 nucleotides were added to a final concentration of 500 µM each. After 10 min, aliquots were removed and added to a 2× Stop buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). Samples were analysed on an 8% denaturing PAGE (19:1 acrylamide: bis acrylamide, 7 M urea, 1X TBE pH= 8.3) for 1.25 h at 400 V, and the gel was exposed on a Storage Phosphor Screen and imaged using a Typhoon PhosphoImager (GE Healthcare).
Quantification of termination and changes in termination
Synthesized RNA bands on the gel image were quantified using ImageJ software (NIH). Each lane from below the rrf termination site (~150 nt) to above the runoff RNA products (263 nt) was converted to a pseudo-densitometer plot using the ImageJ line function and the relative areas of the termination and runoff bands were measured. Termination efficiency (TE) was calculated as the fraction of the termination (term) peak area relative to total of the termination and runoff (term + runoff) peak areas. Fold changes in termination attributable to each NusG (∆T) were determined as the aggregate of changes in the termination rates kb and kt, as defined by von Hippel and Yager (equations (1) and (2))62,63. Multiple algebraic transforms can yield the aggregate fold changes in termination, ∆T, based on the following equations.
$${\rm{TE}}=\frac{{k}_{t}}{{k}_{t}+{k}_{b}}$$
(1)
$${\rm{TE}}={\left[1+{{\rm{e}}}^{-\Delta \Delta {G}^{\ddagger }/-RT}\right]}^{-1},$$
(2)
where ∆∆G‡ is the difference in activation barriers between termination and bypass, which is most directly related to the energies of RNAP–NusG and internal RNAP interactions that govern termination.
$${\Delta \Delta G}^{\ddagger }=-RT\times {\rm{ln}}\left(\left(1/{\rm{TE}}\right)-1\right)$$
(3)
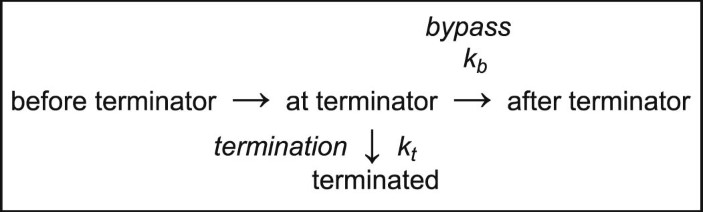
(equation (2) rearranged).
$$\Delta T={{\rm{e}}}^{\left({\Delta \Delta G}_{2}^{\ddagger }-{\Delta \Delta G}_{1}^{\ddagger }\right)}$$
(4)
(fold change in aggregate termination rates for two conditions, 1 and 2).
$$\Delta T=\frac{\left(\frac{1}{{{\rm{TE}}}_{2}}\right)-1}{\left(\frac{1}{{{\rm{TE}}}_{1}}\right)-1}$$
(5)
(alternative calculation derived from equation (1) assuming NusG only affects kb).
Calculating ∆T using either the combinations of equations (3) and (4) or using equation (5) gives the same results because the ∆T is the same whether conditions differ by aggregate effects on both kb and kt or an effect on only one of them. We calculate ∆T using these approaches rather than the simple difference in energies of activation (\(\Delta \Delta {G}_{2}^{\ddagger }-\Delta \Delta {G}_{1}^{\ddagger }\)) because it allows a clearer graphical depiction of effects without changing the results. Errors in ∆T were calculated using a two-sided, unpaired t-test with no assumptions on variance.
Electrophoretic mobility shift assay
RNAP–NusG complexes were assembled and run on an electrophoretic mobility shift assay to test proper binding of all mutant NusGs. Core RNAP (200 nM) was incubated with the template strand of elongation scaffold DNA13 (50 nM) for 15 min at room temperature. Next, the complex was incubated with the complementary non-template strand (50 nM) for 15 min at room temperature. Finally, the complex was incubated with 1 µM wild-type NusG, N65H NusG, R124L NusG, or N125S NusG for 10 min at room temperature. All complexes were assembled in the following transcription buffer: 20 mM Tris, 25 mM potassium glutamate, 10 mM magnesium acetate, 1 mM DTT, 5 µg ml−1 BSA. Samples were immediately loaded and run on a native PAGE (4.5% acrylamide:bis solution 37.5:1, 4% glycerol, 1× TBE) for 1 h at 15 mA. The gel was run at 4 °C. The gel was first stained with GelRed (Biotium) followed by Coomassie blue for visualization of DNA and protein respectively.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
[ad_2]
Source Article Link

